Hi there!
It’s been a long time since I last blogged, but that is because I’ve been swimming round in data, which has incidentally led to the findings that were published in this paper , which I will describe in this post.
HPV and the link to cancer.
HPV (Human Papillomaviruses) consist of a family of viruses that infect keratinocytes (skin cells) that line the outside of the body and the inner cavities – some of them just cause warts (and genital warts) but some of them are capable of driving the formation of cancer. These types, which are called “High-risk” strains, are the ones that are targeted for prevention by HPV vaccines.
High-risk HPV strains differ from low-risk strains in terms of cancer-causing ability because of proteins they make during their life cycle. Cells need to be actively dividing to permit HPV replication and in order to do this, the virus uses two proteins, called E6 and E7 , to block and degrade two proteins in human cells, called TP53 and pRb, which are two potent tumour suppressors (genes that prevent tumour formation).
Normally, E6 and E7 are only active for a brief while during the virus’ life cycle, which culminates in the production of more viruses that restart the cycle all over again, but before HPV driven cancers form something very strange happens; by complete accident the viral genome gets inserted and integrated into human DNA in infected cells, or infected cells get locked into a state where E6 and E7 are produced all the time. Suddenly you’ve got cells with TP53 and pRb off all the time, leaving behind cells that can grow abnormally. We see this when women have cervical scrapings looked at, and see “dysplastic” cells that have grown clumpy and abnormal.

However, these dysplastic cells are not cancerous – and haven’t acquired all the hallmarks of cancer. For this to happen there need to be additional changes to the DNA sequence (Mutations) of the genes in dysplastic cells that can confer those properties. Well known examples of things that cause mutations include tobacco smoke; for quite a while it had been an open question as to where HPV-driven tumours got their mutations from.
Suspicions are aroused: could the APOBEC family of proteins be making these mutations?
One of my major research interests is to see what genes are expressed more and what genes are turned off in HPV driven cancers, and when defining a signature for these tumours I compared them to normal tissue and HPV negative tumours that arise in the same tissue (while cervical cancers usually all tend to be HPV-driven, there are head and neck cancers caused by HPV and those caused by chronic tobacco and alcohol exposure) and one of the genes that I found expressed at high levels in HPV-positive tumours was APOBEC3B.
APOBEC3B is one of many proteins of the APOBEC cytosine deaminases family. These act either on RNA or DNA when it is a single stranded state, and take part in the body’s immune response against viruses by messing up the RNA/DNA from the viruses. They work by changing cytosines, one of the four bases that make up DNA to uracil (a base that is normally only found in RNA) which then gets converted to a thymine or a guanine (two other bases that make up DNA); so if you get lots of these changes in viral DNA you fundamentally break them so they can’t do any of the things they usually do, and it had been known for a while that you could find HPV with messed up DNA in precancerous lesions with patterns of change associated with APOBEC proteins.
This led us to wonder if APOBEC proteins could end up accidentally changing human DNA just like it would change viral DNA and therefore generate the necessary DNA sequence changes to cause cancer; and at the same time we started wondering that a couple of papers came out showing that there were human cancers in which mutations looked like they were being generated by APOBEC enzymes, very likely APOBEC3B (We could tell it was likely APOBEC 3B because it is known to change cytosines that are preceded by a thymine and followed by guanine or adenine or thymine, so if the sequence was TCA or TCG or TCT it would be converted to TGA/TTA or TGG/TTG or TTT/TGT ). There is an alternative process that can also generate TCG->TGG/TTG mutations, so in order to specifically measure APOBEC activity we ended up using the others, which we referred to in the paper as TCW to TKW (TCW->TKW, where K = G or T and W = A or T).
Those previous papers also noted that cervical cancers had lots of mutations that showed the APOBEC signature, but the question remained – was this down to it being the cervix? or was it down to these tumours being HPV+? We decided to take a look in head and neck cancers as well where we could compare HPV+ and HPV- tumours that arose in similar tissues to see if there was truly an association with HPV, and hence we did the work reported in the paper…
HPV positive tumours have a vastly higher fraction of mutations belonging to the APOBEC signature.
First, we ended up looking at levels of APOBEC mutagenesis and how much of all the mutations in tumours were attributable to them using publicly available data for 40 HPV+ head and neck tumours and 253 HPV- head and neck tumours. To do this we used multiple approaches – including looking at TCW->TKW mutations and also trying to break down all the mutations we see in these tumours into patterns of mutations, as was done by these people at the Sanger Institute , and also looking at enrichment for the TCW->TKW mutation pattern locally. All the approaches we used showed the same thing – HPV+ tumours had a vastly higher proportion of mutations most likely caused by APOBEC enzymes.
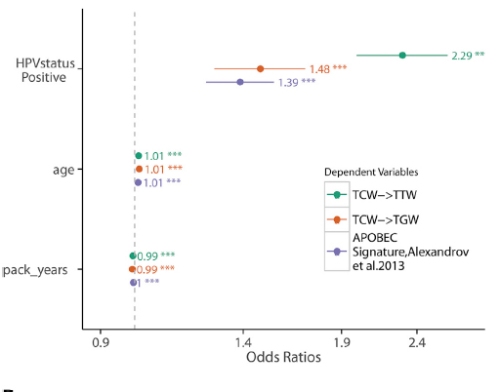
Multiple measures of APOBEC activity showed a strong association with HPV status but not age or smoking; APOBEC, age and smoking were the three processes we identified as driving the signatures using the Sanger Institute’s approach. The more the numbers are shifted to the right the stronger the association with the factor listed on the left.
We found signatures previously associated with APOBEC, smoking and age, and showed that APOBEC activity was not associated with the latter two, which was as expected. Having identified an association with HPV driven tumours we wanted to know if this was a general antiviral response or something HPV specific…so we took a look at patterns of mutations in liver cancers caused by hepatitis B and C viruses and found no evidence for APOBEC mediated mutations being significantly enriched in these tumours.
Of drivers and passengers
Most tumours have hundreds and thousands of mutation, but only a few actively contribute to the acquisition and maintenance of the hallmarks of cancer. So, having initially identified high proportions of APOBEC-mediated mutations in HPV driven cancers when looking across the exome (all protein coding genes in general) we decided to ask if the enrichment we saw in all genes was also maintained when we restricted our searching to genes known previously to drive cancer or those that share features associated with drivers, like occurring at a frequency greater than expected by chance. Our analyses confirmed that APOBEC-mediated mutations were again enriched in the HPV+ head and neck, and cervical cancers compared to the HPV- HNSCs.
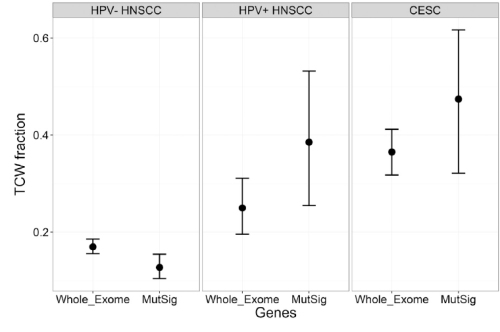
Differences between HPV negative HNSCC and HPV+ tumours (HNSCC and Cervical cancer) are maintained when looking at all protein-coding genes (whole exome) and likely driver mutations (MutSig).
Then we went on to look at which driver genes happened to be most mutated by APOBEC proteins, and found a gene called PIK3CA (one of the components of a protein complex called PI3 kinase) towards the very top of the list. PIK3CA has previously been reported as being vital to the sustenance of many HPV positive tumours in particular and head and neck cancers in general, and drugs are being developed to target it. Interestingly, we observed that in the HPV+ tumours 22/25 PIK3CA mutations recorded were of the APOBEC type, while this wasn’t the case for the HPV negative tumours.
This then led to yet another question – can the levels of APOBEC activity explain a preference for APOBEC mutations in HPV-positive tumours? Now for driver genes there are two things that may govern what kinds of mutations we see – how much of a growth advantage a mutation in a driver gene gives that cell and the mutation itself. My supervisor, Tim Fenton, who worked on PI3 kinases previously, knew that there were two regions in PI3 kinase amongst which mutations regularly occurred (one or the other) and then realised that one of them contained a TCW sequence that APOBEC proteins could act on while the other one did not.
The PIK3CA gene makes a protein called p110-alpha, and proteins have different distinct elements in their structure, called domains. One region, called the helical domain, is often mutated at two TCW sequences while the other region, called a kinase domain, is not, and both mutations confer similar growth advantage, and if you look across multiple tumour types, overall you tend to see a 50-50 split between the two. This enabled us to account for growth advantage and directly see if APOBEC activity, which we had already measured by looking at all protein-coding genes, and a preference for APOBEC-induced mutations in the helical domain, were linked.
Since PIK3CA is mutated in multiple types of cancers, I was able to grab some data from The Cancer Genome Atlas project and measure how strongly there was a skew towards acquiring helical domain mutations compared to the kinase domain mutations and just look at what APOBEC activity looked like in each of those types of tumours. The results were quite robust – the higher the APOBEC activity in a cancer type, the stronger the preference for helical domain mutations compared to kinase domain mutations.
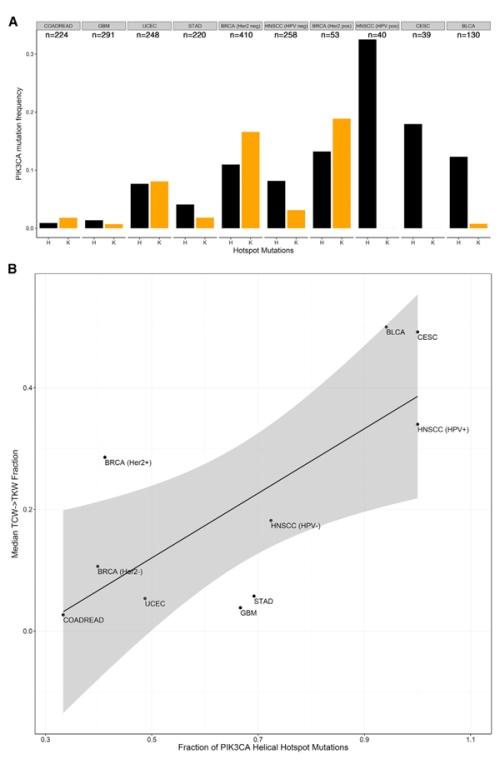
Figure 3. A – as you move from left to right (tumour types are arranged from left to right based on median APOBEC activity), you see helical domain mutations (black bars) become strongly preferred compared to kinase domain mutations (yellow bars). B – plotting the median TCW->TKW fraction (APOBEC activity) against the proportion of PIK3CA mutations that are helical hotspot mutations shows a strong correlation.
So yeah, people had been wondering why in bladder cancers, for example, you saw such a strong preference for helical hotspot mutations – we basically addressed that long-standing question with these analyses.
Explanatory factors
So the one other thing we did was to look at what might be driving this process, and surprisingly we found no correlation between how much E6 and E7 was being expressed in these tumours and APOBEC activity, or for that matter between APOBEC3B gene expression and APOBEC activity, and did find a strong link with how many mutations in total these tumours had. The work has led us to hypothesize it may be something like DNA damage induced by HPV, that generates the substrate for APOBEC3B to act upon, that drives the process.
Conclusion
Our work suggests that HPV positive tumours evolve in a trajectory where they incorporate HPV DNA into their own, leading to sustained E6/E7 expression, followed by APOBEC activity until a driver mutation occurs, after which clones expand and show the APOBEC signature when their DNA is sequenced while in HPV negative HNSCC smoking and alcohol do this job, and if PIK3CA is the gene mutated the HPV positive tumours tend to have helical domain hotspot mutations because APOBEC proteins are responsible for them…
Additional stuff
The journal did a Q&A that expands on some of the work in the paper, and you may find it here .
There is a press release from UCL here.