It is our inalienable heritage. It is humanity’s common thread
– Sir John Sulston.
The story I am going to tell you here is one that I rate extremely significant in the history of our species. It enabled us to identify ourselves at the molecular level, and also established a genetic link to the rest of the biosphere by allowing us to peer into the similarities and the differences we share with the rest of the living world.
It is a story of public co-operation on a scale that is not usually seen, it is a story of the ethical fight to keep the data from the human genome project in the public domain as opposed to being the property of corporate owners who might have very well tried to monopolize genes and what they do, it is a story of scientific prowess and technological achievement, finally it is a story of where we come from and where we are headed, and it tells of great promises that may be fulfilled in our fight against disease & death and greatly help us in our pursuit of good health. It promises to shine a light on the molecular mechanisms of development, of what makes us who we are, and what goes wrong when we fall sick. I think it is one of the most marvellous stories one could perhaps narrate insofar human scientific achievement is concerned.
The Thread of Life – DNA.
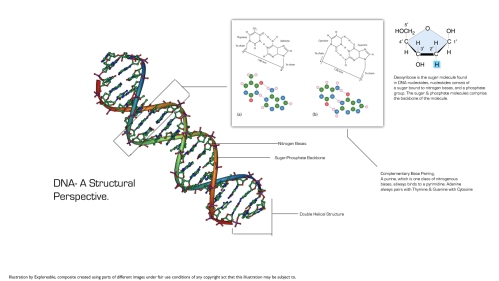
Before we move on to the rest of the article, I feel an introduction to DNA is warranted. DNA is a polymer of nucleic acids that is found in most organisms in a double stranded configuration. DNA is a molecule that acts as a template for the body to make proteins, and organisms are made out of proteins or other molecules which are acted upon by proteins. DNA also has regions which regulate when genes are turned on and off, and these enable things such as signalling and feedback to be introduced. Despite being a bog-standard polymer, DNA has the ability to orchestrate all the complex chemistry that is essential to turn single cells that carry a sufficiently complex genome to extremely complex multicellular organisms. Everything that we are, except whatever behaviours we may learn due to the environment, is down to DNA and processes that act on DNA.
I will be writing more on how this transition from genes to phenes happens in future posts, but until then it may be useful to learn what the structure of DNA is like and what implications this has. DNA in most organisms is made up of two strands wrapped around each other in a double helix. I say most organisms because some viruses, for example, have single stranded DNA.
DNA basically consists of two antiparallel strands wound into a helix, these strands are made of a sugar molecule called deoxyribose and a phosphate molecule. They are bound to nitrogenous bases of two types, namely purines and pyrimidines. A purine always binds to a pyrimidine, i.e Adenine always binds to thymine and Guanine always binds to Cytosine, we call this complementary base pairing. This is extremely relevant to how DNA works… since it can explain how descendent cells can acquire their DNA, i.e, either strand can be used as a template to produce a new double stranded copy of DNA. It can also explain how sequence can determine the sequence of other molecules that are produced using DNA as a template, using processes such as transcription and translation. You may want to read articles that I’ve written on the blog that deal with the Central Dogma of Molecular Biology and The Elucidation of the Genetic Code on this very blog if you feel like taking a little detour at this juncture, for we are just getting started. You may also find this interactive exercise to be useful in learning about DNA structure and base pairing to be an interesting thing to do.
A little experiment you can do
Sure, what better way to get you involved in knowing about the project than to actually make it possible for you to actually carry out one of the key steps involved in the HGP, namely the isolation of DNA? While the HGP used human DNA, for purposes of our little experiment, many other organisms will do, and I suggest using plant material for this.
The following protocol may be carried out.
[1] Grind up some peas or onions, around 50 grams, in around 30-40 ml of warm water with 1-2 tablespoons of salt.
[2] Add some detergent to the mixture to lyse cells from the paste and to break down proteins. Filter this.
[3] To the filtrate, add alcohol drop by drop until you see white fibrous clumps moving into the alcohol layer, which is the upper layer.
[4] You have a DNA sample.
It must be noted that this process is very similar to what happens in the case of research grade projects involving DNA extraction, in fact, the chemistry is much the same! You may also carry out an exercise in isolating DNA, virtually, here Go ahead and give it a try, you know you want to 😉
What did the Human Genome Project Entail & What technological advances were critical ?
It basically entailed a large scale, collaborative sequencing effort to completely sequence the haploid genomes (either eggs or sperm) of genome donors who were anonymous. This effort was helped greatly by the development of high quality, rapid sequencing methods that allowed genome fragments to be sequenced and then put in place by the algorithmic arrangement of overlapping ends to produce a continuous sequence.
The sequencing technology that was used in the HGP was an automated form of Sanger’s Chain Termination Method, for which Fred Sanger won a second Nobel Prize in Chemistry for coming up with this. Automation enabled sequencing studies to handle ever larger genomes, and there people could map genomes both on a whole genome basis and on a fragment based basis.
The major technological platforms that were needed to successfully complete the HGP were improved and developed by working on the genomes of other organisms, ranging from viruses with cute little genomes to organisms of much more complex organisms such as the worm C.elegans (which by itself has won people who studied facets of it four Nobel prizes) and then Saccharomyces cerevisiae (Baker’s yeast, the stuff that brews yer beer). Sequencing the Drosophila melanogaster genome was one of the first projects to utilize the Whole Genome Shotgun method developed by Celera Genomics (Craig Venter’s company).
How Sanger Sequencing Works

The idea here is that once you have single stranded DNA with a starting primer sequence, DNA polymerase will extend it ( this is the same principle used in the extension step of PCR, about which I have written on this blog before), in Sanger sequencing, you label a form of nucleotide that binds complementarily to the single stranded DNA template but does not allow DNA polymerase to extend it further, hence terminating the chain and add this to the mixture. After sorting these chains by size using electrophoresis , we can just read the sequence of terminal bases using the fluorescent labels that they have been treated with, and ta-da, we can read the sequence.
Here is a video that explains the concept of Sanger Chain Termination Sequence to you.
Approaches to Genome Sequencing.
In this section, I describe the two predominant approaches that HGP teams used to organize samples and assemble data from sequencing those samples into genome draft sequences.
Hierarchical Genome Sequencing
This method was implemented by the Publicly funded collaboration. The difference between this and the private Celera Genomics project is apparent in sample preparation and assembly.

In Hierarchical Shotgun Sequencing…
[1] Markers for regions of the genomes are identified.
[2] The genome is split into fragments using restriction/cutting enzymes that contain a known marker.
[3] These fragments are cloned in bacteria using bacterial plasmids, we call these constructs BACs (Bacterial Artificial Chromosomes)
[4] These fragments are individually sequenced using automated Sanger sequencing.
[5] Assembly of the genome is done on the basis of prior knowledge of the markers used to localize sequenced fragments to their genomic location. A computer stitches the sequences up using the markers as a reference guide.
Whole Genome Shotgun Sequencing
This method was employed by Celera Genomics, which was a private entity that was trying to monopolise the human genome sequence by patenting it, to do this they had to try and beat the publicly funded project. Whole genome shotgun sequencing was therefore adopted by them.

Here,
[1] A library is generated of random fragments of the human genome using restriction digestion followed by cloning.
[2] These fragments are then sequenced, we call each of these fragments a sequence read.
[3] Overlapping sequences are then used to produce contiguous sequences.
[4] A scaffold is constructed using computationally predicted read pairs.
[5] Contiguous sequences are then computationally assembled together.
Now the thing with Celera Genomics is that they did data integration using both the scaffold method and the marker method using publicly available data, they found, as a result, that hierarchical sequencing was slightly more efficient, but more or less the data could be analysed and integrated using both approaches.
At this juncture, you may want to take a little detour again to read a more detailed explanation of the technology involved on Nature Scitable, which you may find here
Nature of the Donors whose sequences were used
Due to ethical considerations, the donors have remained anonymous, but the approach the public project used was to isolate White Blood Cells from two male and two female donors, mix it all together and to then put these samples through one of the aforementioned sequencing workflows.
The Celera Genomics project used five anonymous donors and samples that were taken from a pool of 21 donors initially. The source tissue for samples again appears to be similar to that in the public project.
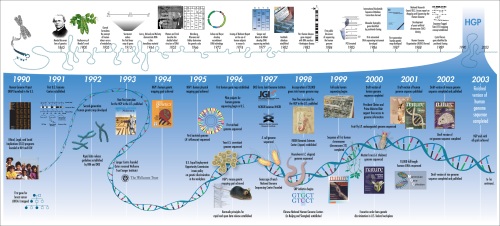
The timeline of the Human Genome Project, and some comments.
[1] Much of the work that made sequencing the human genome possible actually took place something like more than a century ago, starting with the elucidation of the nature of inheritance, subsequent localization of heredity to chromosomes, then confirmation of DNA as the genetic material and finally the discovery of the double helical structure of DNA. This was then followed by discovering how DNA replication took place, how DNA specifies protein sequences and further insights from the role of genes and proteins in development.
[2] Along the way, we learned how variations in genes can contribute to disease, starting with the implication of genes in Huntington’s Chorea and the discovery of the variant gene that causes Duchenne’s Muscular Dystrophy. The development of Sanger sequencing was a landmark.
[3] Sequencing the genomes of several model organisms along the way is very significant in my opinion because it allows us to use genetic data from those organisms in relation to our own so that they can be of import to human studies and biology. We already know of genes such as Pax6 which are universally conserved in the development of animal eyes, for instance. We are able to carry out precise comparison between gene sequences in order to identify what genetic differences are responsible for phenetic differences.
[4] The immense amounts of co-operation seen, with the publicly funded project actively involving large amounts of international partners is an indicator of what could be achieved if only people were willing to look beyond the barriers of nationalism, in my opinion. The fact that we had sequencing centers from North America, Europe & Asia is also an indicator of how important one could hold the common heritage that we all share to be.
[5] The completion of sequencing of the first chromosome in the HGP, namely Chromosome 22 was a significant landmark since it showed it could be done and was only a matter of time.
[6] The announcement of a completed draft in 2003 was a beautiful moment in the history of science, since a thirteen year project had now officially ended with all its goals achieved.
Of Public Affairs and Private Affairs
Now this is a very interesting bit, people had been patenting genes for long but the sequencing of the human genome brought with it its own aspirants for monopoly. Celera wanted to patent the human genome in case it got there first, and charge researchers for the privilege of accessing said data, and to prevent redistribution. They also wanted to release data annually, which could stymy progress.
The public effort, on the other hand, has had a policy of open access to the data it publishes, with new data to be mandatorily published as soon as the compilation of the sequence following sequence was complete, within 24 hours. I have always been one for open access, and I find it tremendously relieving that I do not have to pay to access what is a part of my own molecular heritage, that would seriously suck.
Of spinoffs, benefits & implications.
[1] The completion of the draft enabled variation studies to be carried out, such as HapMap, which documents single nucleotide variations (or point mutations/substitutions) which have diagnostic and prognostic value.
[2] The availability of a reference sequence enabled the study of the functioning of normal vs mutant genes and in deriving a working understanding of how aberrant gene function may be linked with diseases. This also encouraged the development of diagnostic markers for diseases, a case in point being the detection of a test for BRCA1 and BRCA2 variants which are good markers for susceptibility to familial breast cancers.
[3] Having reference sequences available along with variation data can help in the analysis of intragenic variation and how evolution might be shaping the genome, for instance, it can also optimize the development of gene silencing techniques which may enable us to not only study gene function, but to utilize the same to develop therapeutic strategies to combat those genetic disorders.
[4] In diseases like cancer, it facilitates analysis of alleles mutated or implicated in disease progression, this gives us understanding of pathogenesis at the molecular level and can inform the search for drug targets that can be exploited to treat the disease and the drugs to actually attack those targets.
[5] Projects like ENCODE, which are involved in the identification of all expressed DNA elements, not just the 25000 or so protein expressing sequences, will help us to further our understanding of how genes work in concert to bring phenes into existence and as a consequence how our genomes make us who we are with the exception of external environmentally shaped traits.
[6] The availability of the human genome sequence can further molecular diagnostics involving PCR for instance by enabling PCR primer design that does not cross react with human transcripts, which means that pathogen DNA/RNA can be reliably amplified and identified for diagnostic purposes.
[7] The development of unique microarray probes that has now made large scale array based expression studies possible, improving our understanding of the pathology of diseases, is also something that owes its origins to this project.
[8] As gene sequencing technology improves, and we are able to sequence more and more for less and less money and to it quicker, we should start seeing masses of genetic data emerging that can be seamlessly integrated into databases that deal with the human genome and variation therein, and this will not only benefit researchers as they learn more and more from such data but will also help the people who have their genome sequenced by helping them make use of information gleaned from what researchers are looking at.
There is already technology being developed that could make genetic sequencing dizzyingly fast and efficient, including for instance Nanopore sequencing where nanotechnology is used to drive sequencing.
Another such large scale sequencing technology under development is the Helicos synthesizer, if it works well it could be a brilliant addition to the toolkit molecular geneticists have.
[9] The project helped to establish that all humans share a common heritage, this is summed up in the very first lines by a quote by Dr.John Sulston, who I am aware was the director of the Wellcome Trust’s Sanger Center when the project was running. So this means anybody who starts to discriminate amongst people based on superficial variation can screw themselves, and IMO *should* screw themselves.
[10] It also raised ethical concerns about genetic privacy and what insurance companies could treat people with susceptibility to certain disorders, in the US of A, at least, there was a ban against federal genetic discrimination introduced in about 2000.
It remains to be seen how effective legislation against genetic discrimination is, but I fervently hope that this doesn’t become a problem.
[11] On a much more individualistic level, it may help parents who are carriers of genetic diseases to choose to have children by checking through genetic testing if the embryo is disease free.
Delving into the Details – More you can do
Books
[1] The Common Thread, Sir John Sulston & Georgina Ferry, Joseph Henry Press.
I’ve had the pleasure of reading this nearly autobiographical book, and it is a brilliant, gripping account of what happened during the HGP, written by someone who was at the forefront of research involved in the project : Sir Sulston was the director at the Sanger Centre.
The Google Books entry for this can be found here.
[2] Genome : The autobiography of a species in 23 chapters by Matt Ridley.
This is a popular science treatment of the Human Genome & the HGP for laymen, you may find this a useful read if you are looking for a platform to delve into exploring more of this unique scientific milestone. You may find the apposite Amazon Page here
[3] Our Posthuman Future, Francis Fukuyama, Picador Press.
This book is not per se about the Human Genome, but it covers issues like the ethics of biotechnology and genetic engineering and other technologies. It could be useful for anybody trying to understand where the ethics regarding biotechnology policymaking lie and what the implications are.
Television.
[1] The Gene Code – A BBC4 Documentary written and presented by Dr.Adam Rutherford, this two part programme introduces you to DNA and then takes you through the Human Genome Project and its implications, but be warned that some of the things mentioned therein are less than academically rigorous, as in the failure to differentiate between junk and ncDNA, for instance.
[2] The Incredible Human Journey – A BBC Documentary presented by Dr.Alice Roberts, this five part programme traces the routes humanity took out of Africa and how our species spread all over the world. It is something that encapsulates the commonality of our heritage, both cultural and genetic very well.
Websites
You may want to,
[1] Access a Nature Scitable book on the Human Genome Project here
[2] Access a Nature Scitable book on Genomes and their links to diseases
[3] How do we map genes and link genes to phenes? Find out more by browsing this nature Scitable book titled Gene Mapping: Then and Now.
[4] If you want to learn how to browse the human genome, look for genes and then take a tour into the world of genomic data, you may find this tutorial, which deals with the use of a software portal called the UCSC Genome Browser, which is very useful when it comes to perusing genomic data. If you want to go through the genome, look at genes and what they do, this is one very versatile tool to facilitate that.
[5] Read the official Department of Energy’s (the primary financier of the HGP in the USA) HGP Portal here
[6] Visit the Wellcome Trust’s beautiful educational resource site for the HGP, called YourGenome.org You can find animations, explanations & activities listed on this site which may make learning about the HGP, and genes and genomes a very interesting proposition.
That is all (!) from me as far as this particular post is concerned. I hope you had a happy time reading & stayed awake throughout, if not I expect your gratitude for helping you to doze off 😛 .
– Ankur “Exploreable” Chakravarthy.

Hi there, just ran into this web site from reddit. It is not an article I would normally read, but I loved your thoughts on it. Thanks for creating an article worth reading!
I was just seeking this information for some time. After six hours of continuous Googleing, finally I got it in your website. I wonder what is the lack of Google strategy that do not rank this type of informative web sites in top of the list. Generally the top websites are full of garbage.
Nice..
Pingback: Les estadístiques del genoma | e2013
Pingback: A Bird’s Eye View of Cancer Research… | Exploreable
Pingback: Resum Jornada estadísTICa (2) | e2013